DATA ANALYSIS - BOSTON HOUSING PRICES
Task 1. Loading the data
The first step before proceeding is to load the data, which is stored in the file housing.csv.
To do this, we are going to use the Pandas library.
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
data = pd.read_csv("housing.csv",sep='\s+',names=["crim", "zn", "indus","chas","nox","rm","age","dis","rad","tax","ptratio","black","lstat","medv"])
data
| crim | zn | indus | chas | nox | rm | age | dis | rad | tax | ptratio | black | lstat | medv | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.00632 | 18.0 | 2.31 | 0 | 0.538 | 6.575 | 65.2 | 4.0900 | 1 | 296.0 | 15.3 | 396.90 | 4.98 | 24.0 |
| 1 | 0.02731 | 0.0 | 7.07 | 0 | 0.469 | 6.421 | 78.9 | 4.9671 | 2 | 242.0 | 17.8 | 396.90 | 9.14 | 21.6 |
| 2 | 0.02729 | 0.0 | 7.07 | 0 | 0.469 | 7.185 | 61.1 | 4.9671 | 2 | 242.0 | 17.8 | 392.83 | 4.03 | 34.7 |
| 3 | 0.03237 | 0.0 | 2.18 | 0 | 0.458 | 6.998 | 45.8 | 6.0622 | 3 | 222.0 | 18.7 | 394.63 | 2.94 | 33.4 |
| 4 | 0.06905 | 0.0 | 2.18 | 0 | 0.458 | 7.147 | 54.2 | 6.0622 | 3 | 222.0 | 18.7 | 396.90 | 5.33 | 36.2 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 501 | 0.06263 | 0.0 | 11.93 | 0 | 0.573 | 6.593 | 69.1 | 2.4786 | 1 | 273.0 | 21.0 | 391.99 | 9.67 | 22.4 |
| 502 | 0.04527 | 0.0 | 11.93 | 0 | 0.573 | 6.120 | 76.7 | 2.2875 | 1 | 273.0 | 21.0 | 396.90 | 9.08 | 20.6 |
| 503 | 0.06076 | 0.0 | 11.93 | 0 | 0.573 | 6.976 | 91.0 | 2.1675 | 1 | 273.0 | 21.0 | 396.90 | 5.64 | 23.9 |
| 504 | 0.10959 | 0.0 | 11.93 | 0 | 0.573 | 6.794 | 89.3 | 2.3889 | 1 | 273.0 | 21.0 | 393.45 | 6.48 | 22.0 |
| 505 | 0.04741 | 0.0 | 11.93 | 0 | 0.573 | 6.030 | 80.8 | 2.5050 | 1 | 273.0 | 21.0 | 396.90 | 7.88 | 11.9 |
506 rows × 14 columns
Task 2. Exploratory analysis
# DATA DIMENSIONS
print("Dimensions of the data:",data.shape)
print(data.shape[0], "instances.")
print(data.shape[1], "attributes.")
Dimensions of the data: (506, 14)
506 instances.
14 attributes.
data.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 506 entries, 0 to 505
Data columns (total 14 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 crim 506 non-null float64
1 zn 506 non-null float64
2 indus 506 non-null float64
3 chas 506 non-null int64
4 nox 506 non-null float64
5 rm 506 non-null float64
6 age 506 non-null float64
7 dis 506 non-null float64
8 rad 506 non-null int64
9 tax 506 non-null float64
10 ptratio 506 non-null float64
11 black 506 non-null float64
12 lstat 506 non-null float64
13 medv 506 non-null float64
dtypes: float64(12), int64(2)
memory usage: 55.5 KB
Impressions: it can be seen that there are no missing null gaps in the attribute values as there are 506 instances and each of the attributes contains 506 values, all columns are numeric (including the class, being a regression problem).
data.describe()
| crim | zn | indus | chas | nox | rm | age | dis | rad | tax | ptratio | black | lstat | medv | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 506.000000 | 506.000000 | 506.000000 | 506.000000 | 506.000000 | 506.000000 | 506.000000 | 506.000000 | 506.000000 | 506.000000 | 506.000000 | 506.000000 | 506.000000 | 506.000000 |
| mean | 3.613524 | 11.363636 | 11.136779 | 0.069170 | 0.554695 | 6.284634 | 68.574901 | 3.795043 | 9.549407 | 408.237154 | 18.455534 | 356.674032 | 12.653063 | 22.532806 |
| std | 8.601545 | 23.322453 | 6.860353 | 0.253994 | 0.115878 | 0.702617 | 28.148861 | 2.105710 | 8.707259 | 168.537116 | 2.164946 | 91.294864 | 7.141062 | 9.197104 |
| min | 0.006320 | 0.000000 | 0.460000 | 0.000000 | 0.385000 | 3.561000 | 2.900000 | 1.129600 | 1.000000 | 187.000000 | 12.600000 | 0.320000 | 1.730000 | 5.000000 |
| 25% | 0.082045 | 0.000000 | 5.190000 | 0.000000 | 0.449000 | 5.885500 | 45.025000 | 2.100175 | 4.000000 | 279.000000 | 17.400000 | 375.377500 | 6.950000 | 17.025000 |
| 50% | 0.256510 | 0.000000 | 9.690000 | 0.000000 | 0.538000 | 6.208500 | 77.500000 | 3.207450 | 5.000000 | 330.000000 | 19.050000 | 391.440000 | 11.360000 | 21.200000 |
| 75% | 3.677082 | 12.500000 | 18.100000 | 0.000000 | 0.624000 | 6.623500 | 94.075000 | 5.188425 | 24.000000 | 666.000000 | 20.200000 | 396.225000 | 16.955000 | 25.000000 |
| max | 88.976200 | 100.000000 | 27.740000 | 1.000000 | 0.871000 | 8.780000 | 100.000000 | 12.126500 | 24.000000 | 711.000000 | 22.000000 | 396.900000 | 37.970000 | 50.000000 |
data.hist(column="medv",bins=10)
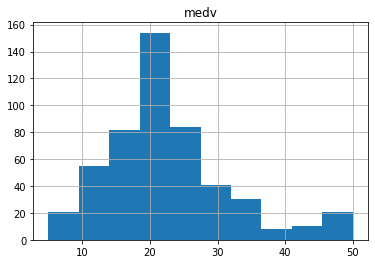
Impressions: roughly following a normal distribution with a slight tail to the right, the median value of the most common inhabited dwellings is around $20,000.
for col in data.columns:
fig,ax = plt.subplots(1,1,figsize=(5,1))
sns.boxplot(x=data[col],showmeans=True)
plt.show()


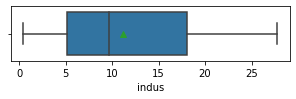

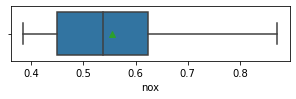
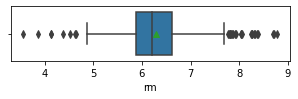
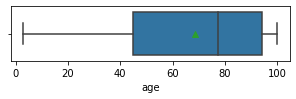
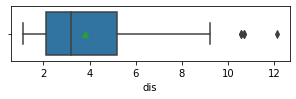


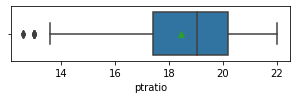

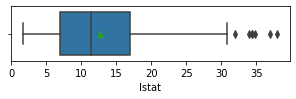
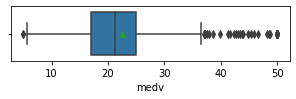
Impressions: it seems that there are variables that have outliers, such as crim, zn, dis, ptratio, lstat, rm, black and the class to predict medv itself. It might be a good idea to remove outliers and then predict on the filtered data.
plt.figure(figsize=(20, 10))
sns.heatmap(data.corr().abs(), cmap='Greens', annot=True)
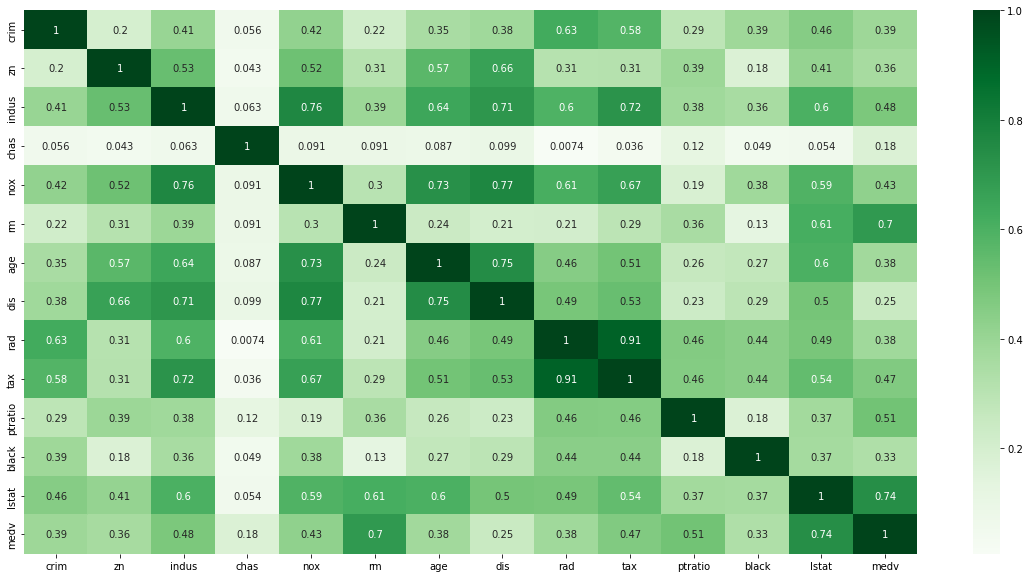
Impressions: Through this correlation matrix we observe several things:
- There is a strong correlation between rad and tax variables.
- The variables that correlate best with medv are rm and lstat (correlation coefficient >= 0.7).
- The variables that correlate worst with medv are dis, black, crim, zn, age, rad. It may be interesting to try removing some of these variables later in the proposed model improvement to see how they actually influence the model (correlation does not imply causation).
sns.set_theme(style="ticks")
sns.scatterplot(data=data,x="crim",y="medv")
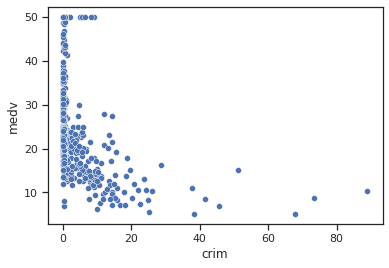
Impressions: It appears that when the per capita crime rate is lower the median house value includes all types of values, high and low, while when the rate is increasing the median house value starts to decrease.
sns.scatterplot(data=data,x="zn",y="medv")
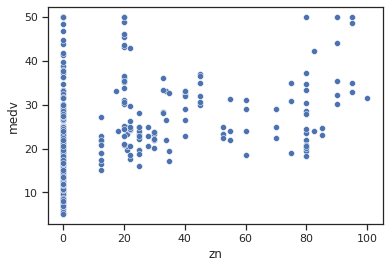
Impressions: some possible relationship between zn and medv is observed when the proportion of residential area in areas larger than 25,000 sq. ft. is non-zero, with slightly higher average housing values observed when the proportion is higher.
sns.scatterplot(data=data,x="indus",y="medv")
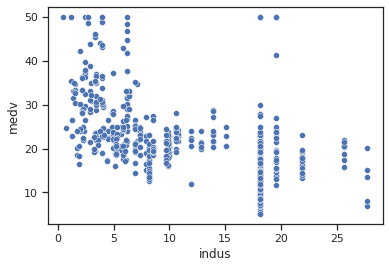
Impressions: The relationship between indus and medv is not very clear, but a slightly decreasing trend is observed where a line could be drawn. It seems that when there is a higher proportion of business extension (excl. retail trade) in the city the median value of inhabited dwellings is lower, while if the proportion is low the median value is rather higher.
sns.scatterplot(data=data,x="nox",y="medv")
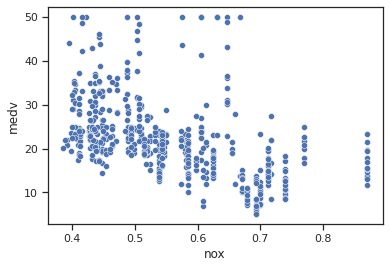
Impressions: It appears that when the concentration of nitrogen oxides is higher the median dwelling value is lower, while lower concentration values have higher median dwelling values.
sns.scatterplot(data=data,x="rm",y="medv")
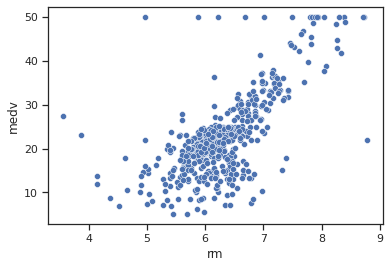
Impressions: There is a strong relationship between the average number of rooms per dwelling and the average value of the dwelling, which is almost a linear relationship (correlation coefficient of 0.7). It seems that the higher the average number of rooms, the higher the average value of the dwelling.
sns.scatterplot(data=data,x="age",y="medv")
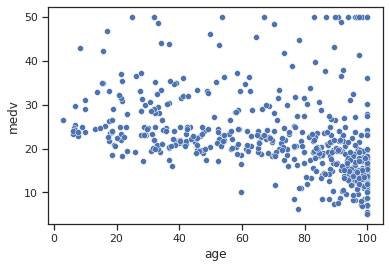
Impressions: where the proportion of pre-1940 occupied dwellings is high, there appear to be more lower average house values, although high values are also included, and as the proportion decreases, the average house value increases.
sns.scatterplot(data=data,x="dis",y="medv")
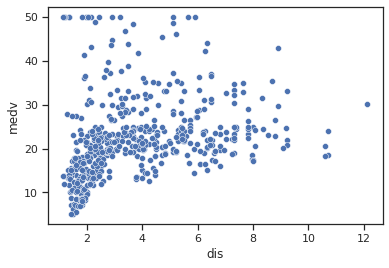
Impressions: When the weighted distance to five employment centres in Boston is low it appears that the median house value is lower overall (although a smaller number of high values are also included) and that as the distance increases the median house value increases.
sns.scatterplot(data=data,x="rad",y="medv")
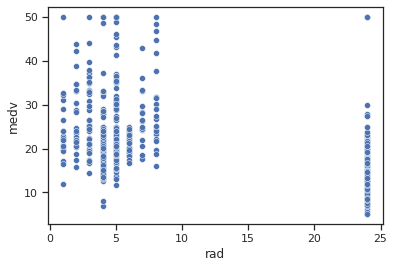
Impressions: it appears that when the radial motorway accessibility index is low there are higher average housing values than when the index is high.
sns.scatterplot(data=data,x="tax",y="medv")
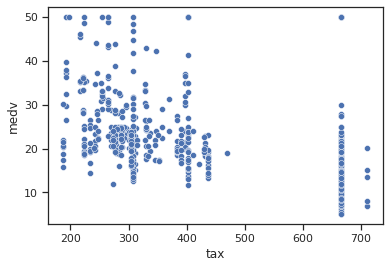
Impressions: it appears that when the property value tax per $10,000 is lower there are higher median housing values, whereas if the tax is high, lower housing values are found.
sns.scatterplot(data=data,x="ptratio",y="medv")
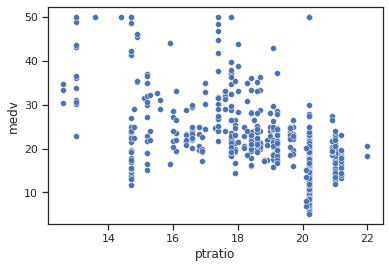
Impressions: it seems that when the pupil-teacher ratio in the city is higher there are lower average housing values, while if the ratio is low, higher average housing values are found.
sns.scatterplot(data=data,x="black",y="medv")
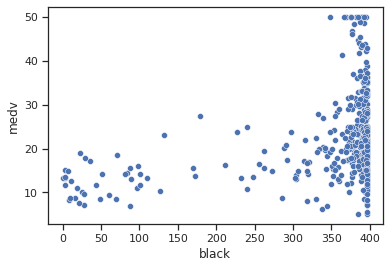
Impressions: no linear relationship is observed, it seems that higher values are found with higher average housing values.
sns.scatterplot(data=data,x="lstat",y="medv")
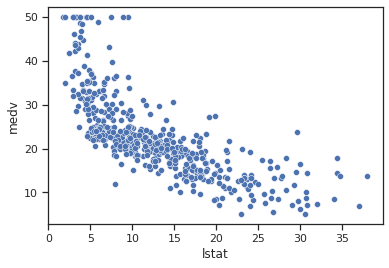
Impressions: There is a good linear relationship between the percentage of the population that is lower class and the median house value. It seems that the higher the percentage of the population that is lower class, the lower the median house value.
Task 3. Linear regression model
from sklearn.linear_model import LinearRegression
from math import sqrt
X=data[["crim", "zn", "indus","chas","nox","rm","age","dis","rad","tax","ptratio","black","lstat"]]
y=data["medv"]
reg = LinearRegression().fit(X, y)
y_pred=reg.predict(X)
print(reg.coef_)
print(reg.intercept_)
[-1.08011358e-01 4.64204584e-02 2.05586264e-02 2.68673382e+00
-1.77666112e+01 3.80986521e+00 6.92224640e-04 -1.47556685e+00
3.06049479e-01 -1.23345939e-02 -9.52747232e-01 9.31168327e-03
-5.24758378e-01]
36.4594883850902
# R2
from sklearn.metrics import r2_score
r2_score(y,y_pred)
0.7406426641094095
# RMSE
from sklearn.metrics import mean_squared_error
sqrt(mean_squared_error(y, y_pred))
4.679191295697281
Analysis: An acceptable prediction is obtained on the training data, with an R2 coefficient not very close to the optimal value 1 , but also not offering random results.
The root mean square error is slightly high, in line with the R2 coefficient.
Task 4. Improving the linear regression model
Regularization methods
# L1 Lasso
from sklearn import linear_model
reg_lasso = linear_model.Lasso(alpha=1.0)
reg_lasso.fit(X,y)
y_pred_lasso=reg_lasso.predict(X)
reg_lasso.score(X,y)
print("R2:",r2_score(y,y_pred_lasso))
print("RMSE:",sqrt(mean_squared_error(y, y_pred_lasso)))
R2: 0.6825842212709925
RMSE: 5.176494871751199
# L2 Ridge
reg_ridge=linear_model.Ridge(alpha=1.0)
reg_ridge.fit(X,y)
y_pred_ridge=reg_ridge.predict(X)
print("R2:",reg_ridge.score(X,y))
print("RMSE:",sqrt(mean_squared_error(y, y_pred_ridge)))
R2: 0.7388703133867616
RMSE: 4.695151993608747
# L1 and L2 : Elastic net
reg_ElasticNet=linear_model.ElasticNet(alpha=1.0)
reg_ElasticNet.fit(X,y)
y_pred_ElasticNet=reg_ElasticNet.predict(X)
print("R2:",reg_ElasticNet.score(X,y))
print("RMSE:",sqrt(mean_squared_error(y, y_pred_ElasticNet)))
R2: 0.6861018474345025
RMSE: 5.147731803213881
Analysis: no regularisation method has improved the prediction on the training data, which makes sense since these regularisations aim to make the model generalise better, thus being less specific, increasing the error.
Reduction of features
As mentioned in the exploratory analysis, after the observation made in the correlation matrix, we will try to eliminate those variables that do not present a high correlation between them and the variable to be predicted.
These are:
- Crim
- Zn
- Chas
- Age
- Dis
- Rad
- Black
#Dropping variables: crim
X_delete=data[["zn", "indus", "rad","chas", "age","nox","rm","dis","tax","ptratio","black","lstat"]]
y=data["medv"]
reg_delete = LinearRegression().fit(X_delete, y)
y_pred_delete=reg_delete.predict(X_delete)
print("R2:",r2_score(y,y_pred_delete))
print("RMSE:",sqrt(mean_squared_error(y, y_pred_delete)))
R2: 0.7349488253339125
RMSE: 4.730275100243263
#Dropping variables: zn
X_delete=data[["crim", "indus", "rad","chas", "age","nox","rm","dis","tax","ptratio","black","lstat"]]
y=data["medv"]
reg_delete = LinearRegression().fit(X_delete, y)
y_pred_delete=reg_delete.predict(X_delete)
print("R2:",r2_score(y,y_pred_delete))
print("RMSE:",sqrt(mean_squared_error(y, y_pred_delete)))
R2: 0.7346146839915815
RMSE: 4.733255812629868
#Dropping variables: chas
X_delete=data[["crim", "indus", "rad","zn", "age","nox","rm","dis","tax","ptratio","black","lstat"]]
y=data["medv"]
reg_delete = LinearRegression().fit(X_delete, y)
y_pred_delete=reg_delete.predict(X_delete)
print("R2:",r2_score(y,y_pred_delete))
print("RMSE:",sqrt(mean_squared_error(y, y_pred_delete)))
R2: 0.7355165089722999
RMSE: 4.725206759835133
#Dropping variables: age
X_delete=data[["crim", "indus", "rad","chas", "zn","nox","rm","dis","tax","ptratio","black","lstat"]]
y=data["medv"]
reg_delete = LinearRegression().fit(X_delete, y)
y_pred_delete=reg_delete.predict(X_delete)
print("R2:",r2_score(y,y_pred_delete))
print("RMSE:",sqrt(mean_squared_error(y, y_pred_delete)))
R2: 0.7406412165505145
RMSE: 4.679204353734578
#Dropping variables: dis
X_delete=data[["crim", "indus", "rad","chas", "age","nox","rm","zn","tax","ptratio","black","lstat"]]
y=data["medv"]
reg_delete = LinearRegression().fit(X_delete, y)
y_pred_delete=reg_delete.predict(X_delete)
print("R2:",r2_score(y,y_pred_delete))
print("RMSE:",sqrt(mean_squared_error(y, y_pred_delete)))
R2: 0.7117915551680203
RMSE: 4.932588467850775
#Dropping variables: rad
X_delete=data[["crim", "indus", "zn","chas", "age","nox","rm","dis","tax","ptratio","black","lstat"]]
y=data["medv"]
reg_delete = LinearRegression().fit(X_delete, y)
y_pred_delete=reg_delete.predict(X_delete)
print("R2:",r2_score(y,y_pred_delete))
print("RMSE:",sqrt(mean_squared_error(y, y_pred_delete)))
R2: 0.7294255414274199
RMSE: 4.779307031347956
#Dropping variables: black
X_delete=data[["crim", "indus", "rad","chas", "age","nox","rm","dis","tax","ptratio","zn","lstat"]]
y=data["medv"]
reg_delete = LinearRegression().fit(X_delete, y)
y_pred_delete=reg_delete.predict(X_delete)
print("R2:",r2_score(y,y_pred_delete))
print("RMSE:",sqrt(mean_squared_error(y, y_pred_delete)))
R2: 0.7343070437613076
RMSE: 4.735998462783738
#Dropping variables: crim zn chas age dis rad black
X_delete=data[["rm","lstat","ptratio","nox","black"]]
y=data["medv"]
reg_delete = LinearRegression().fit(X_delete, y)
y_pred_delete=reg_delete.predict(X_delete)
print("R2:",r2_score(y,y_pred_delete))
print("RMSE:",sqrt(mean_squared_error(y, y_pred_delete)))
R2: 0.687763171482284
RMSE: 5.1340913976159746
Analysis: after the predictions made by reducing the number of variables, no reduction improves the prediction on the training data. It has been observed that the variable that produces the least error by removing it is age.
Task 5. Generalisation of the linear regression model
import numpy as np
data_shuffle = data.sample(frac=1,random_state=1).reset_index(drop=True)
percentage_split=int(0.7*data_shuffle.shape[0])
train=data_shuffle[:percentage_split]
test=data_shuffle[percentage_split:]
X_train=train[["crim", "zn", "indus","chas","nox","rm","age","dis","rad","tax","ptratio","black","lstat"]]
y_train=train["medv"]
X_test=test[["crim", "zn", "indus","chas","nox","rm","age","dis","rad","tax","ptratio","black","lstat"]]
y_test=test["medv"]
print("Size train data:",len(train),"instances.")
print("Size test data:",len(test),"instances.")
reg_general = LinearRegression().fit(X_train, y_train)
y_pred_general=reg_general.predict(X_test)
print("R2:",r2_score(y_test,y_pred_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_general)))
Size train data: 354 instances.
Size test data: 152 instances.
R2: 0.6776486248371075
RMSE: 5.4268766648382245
Analysis: Attempting to generalise the linear regression model without regularising or eliminating variables results in a larger error than on the Task 3 training data.
Regularisation methods
# L1 Lasso
reg_lasso_general = linear_model.Lasso(alpha=0.005)
reg_lasso_general.fit(X_train,y_train)
y_pred_lasso_general=reg_lasso_general.predict(X_test)
print("R2:",r2_score(y_test,y_pred_lasso_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_lasso_general)))
R2: 0.6781960540319253
RMSE: 5.422266644037676
# L2
reg_ridge_general=linear_model.Ridge(alpha=0.3)
reg_ridge_general.fit(X_train,y_train)
y_pred_ridge_general=reg_ridge_general.predict(X_test)
print("R2:",r2_score(y_test,y_pred_ridge_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_ridge_general)))
R2: 0.6777253734106449
RMSE: 5.426230584397384
# L1 and L2 : Elastic net
reg_ElasticNet_general=linear_model.ElasticNet(alpha=1.0)
reg_ElasticNet_general.fit(X_train,y_train)
y_pred_ElasticNet_general=reg_ElasticNet_general.predict(X_test)
print("R2:",r2_score(y_test,y_pred_ElasticNet_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_ElasticNet_general)))
R2: 0.6607401850484715
RMSE: 5.567386838197249
Analysis: after the use of different regulation methods, it is observed that there is no significant improvement in prediction, there is a slight improvement in prediction using Lasso regularisation. It is noticeable the difference of applying these predictions on training data against test data, where a small improvement is observed.
Reduction of variables
As mentioned in the exploratory analysis, after the observation made in the correlation matrix, we will try to eliminate those variables that do not present a high correlation between them and the variable to be predicted.
These are:
- Crim
- Zn
- Chas
- Age
- Dis
- Rad
- Black
#Dropping variables: crim
X_delete_train=train[["zn", "indus", "rad","chas", "age","nox","rm","dis","tax","ptratio","black","lstat"]]
X_delete_test=test[["zn", "indus","rad","chas","age","nox","rm","dis","tax","ptratio","black","lstat"]]
reg_delete_general = LinearRegression().fit(X_delete_train, y_train)
y_pred_delete_general=reg_delete_general.predict(X_delete_test)
print("R2:",r2_score(y_test,y_pred_delete_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_delete_general)))
R2: 0.6759286063605116
RMSE: 5.441335901433361
#Dropping variables: zn
X_delete_train=train[["crim", "indus", "rad","chas", "age","nox","rm","dis","tax","ptratio","black","lstat"]]
X_delete_test=test[["crim", "indus","rad","chas","age","nox","rm","dis","tax","ptratio","black","lstat"]]
reg_delete_general = LinearRegression().fit(X_delete_train, y_train)
y_pred_delete_general=reg_delete_general.predict(X_delete_test)
print("R2:",r2_score(y_test,y_pred_delete_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_delete_general)))
R2: 0.6673861423192364
RMSE: 5.512585743374873
#Dropping variables: chas
X_delete_train=train[["crim", "indus", "rad","zn", "age","nox","rm","dis","tax","ptratio","black","lstat"]]
X_delete_test=test[["crim", "indus","rad","zn","age","nox","rm","dis","tax","ptratio","black","lstat"]]
reg_delete_general = LinearRegression().fit(X_delete_train, y_train)
y_pred_delete_general=reg_delete_general.predict(X_delete_test)
print("R2:",r2_score(y_test,y_pred_delete_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_delete_general)))
R2: 0.6753815942634213
RMSE: 5.445926281290249
#Dropping variables: age
X_delete_train=train[["zn", "indus", "rad","chas", "crim","nox","rm","dis","tax","ptratio","black","lstat"]]
X_delete_test=test[["zn", "indus","rad","chas","crim","nox","rm","dis","tax","ptratio","black","lstat"]]
reg_delete_general = LinearRegression().fit(X_delete_train, y_train)
y_pred_delete_general=reg_delete_general.predict(X_delete_test)
print("R2:",r2_score(y_test,y_pred_delete_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_delete_general)))
# L2
reg_ridge_general_age=linear_model.Ridge(alpha=0.3)
reg_ridge_general_age.fit(X_delete_train,y_train)
y_pred_ridge_general_age=reg_ridge_general_age.predict(X_delete_test)
print("R2:",r2_score(y_test,y_pred_ridge_general_age))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_ridge_general_age)))
R2: 0.6810237811595643
RMSE: 5.398391048362369
R2: 0.6825063159972143
RMSE: 5.385831140417877
#Dropping variables: dis
X_delete_train=train[["crim", "indus", "rad","zn", "age","nox","rm","chas","tax","ptratio","black","lstat"]]
X_delete_test=test[["crim", "indus","rad","zn","age","nox","rm","chas","tax","ptratio","black","lstat"]]
reg_delete_general = LinearRegression().fit(X_delete_train, y_train)
y_pred_delete_general=reg_delete_general.predict(X_delete_test)
print("R2:",r2_score(y_test,y_pred_delete_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_delete_general)))
R2: 0.6566467185102627
RMSE: 5.6008738256069535
#Dropping variables: rad
X_delete_train=train[["crim", "indus", "dis","zn", "age","nox","rm","chas","tax","ptratio","black","lstat"]]
X_delete_test=test[["crim", "indus","dis","zn","age","nox","rm","chas","tax","ptratio","black","lstat"]]
reg_delete_general = LinearRegression().fit(X_delete_train, y_train)
y_pred_delete_general=reg_delete_general.predict(X_delete_test)
print("R2:",r2_score(y_test,y_pred_delete_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_delete_general)))
R2: 0.665046948073654
RMSE: 5.531936134290245
#Dropping variables: black
X_delete_train=train[["crim", "indus", "dis","zn", "age","nox","rm","chas","tax","ptratio","rad","lstat"]]
X_delete_test=test[["crim", "indus","dis","zn","age","nox","rm","chas","tax","ptratio","rad","lstat"]]
reg_delete_general = LinearRegression().fit(X_delete_train, y_train)
y_pred_delete_general=reg_delete_general.predict(X_delete_test)
print("R2:",r2_score(y_test,y_pred_delete_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_delete_general)))
R2: 0.6743000187926087
RMSE: 5.454991204989396
#Dropping variables: crim zn chas age dis rad black
X_delete_train=train[["rm","lstat","ptratio","nox","black"]]
X_delete_test=test[["rm","lstat","ptratio","nox","black"]]
reg_delete_general = LinearRegression().fit(X_delete_train, y_train)
y_pred_delete_general=reg_delete_general.predict(X_delete_test)
print("R2:",r2_score(y_test,y_pred_delete_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_delete_general)))
R2: 0.6449377831448135
RMSE: 5.6955729415015055
Analysis: generalising with the variable reductions, it is observed that the variable that causes the most noise in the model is age, where a perceptible, although very small, improvement has been achieved. Furthermore, by applying a regularisation method with the model without this variable, there is a very small improvement in generalisation.
Extra task
Three regression models provided by Sklearn will be tested. Specifically:
- Support Vector Regression (SVR).
- Decision Tree Regressor
- Nearest Neighbors Regression
# SUPPORT VECTOR MACHINES - REGRESSION
from sklearn import svm
reg = svm.SVR()
reg.fit(X_train, y_train)
y_pred_general=reg.predict(X_test)
print("R2:",r2_score(y_test,y_pred_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_general)))
R2: 0.14911500540686573
RMSE: 8.816995526071912
Analysis: The generalised prediction on the data using SVR gives very poor results, with an R2 of almost 0 and a higher squared error than with the non-generalised model, it has not been possible to find a hyperplane or a set of them capable of separating the data, it is limited to the linearity of the data.
# REGRESSION TREES
from sklearn import tree
reg = tree.DecisionTreeRegressor()
reg.fit(X_train, y_train)
y_pred_general=reg.predict(X_test)
print("R2:",r2_score(y_test,y_pred_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_general)))
R2: 0.7478565977519767
RMSE: 4.799643627121541
Analysis: Generalised prediction on the data by applying a regression tree gives fairly good results, even improving on the performance obtained with a linear regression model. The ability to capture non-linear relationships between attributes and class has allowed good performance to be obtained.
# NEAREST NEIGHBOURS REGRESSION
from sklearn.neighbors import KNeighborsRegressor
reg = KNeighborsRegressor(n_neighbors=4)
reg.fit(X_train, y_train)
y_pred_general=reg.predict(X_test)
print("R2:",r2_score(y_test,y_pred_general))
print("RMSE:",sqrt(mean_squared_error(y_test, y_pred_general)))
R2: 0.47758482578165273
RMSE: 6.90864822018484
Analysis: Generalised prediction on the data using K-nearest neighbours fails to give good results, resulting in a larger error than the original model. These methods are highly dependent on the quality of the data and their scales, and the data collected are not standardised.